Given recent course work in the online machine learning course via Stanford, in this post, I tie all of the concepts together. The R -code that follows borrows heavily from the Statalgo blog, which has done a tremendous job breaking down the concepts from the course and applying them in R. This is highly redundant to that post, but not quite as sophisticated (i.e. my code isn't nearly as efficient), but it allows me to intuitively extend the previous work I've done.
If you are not going to use a canned routine from R or SAS to estimate a regression, then why not at least apply the matrix inversions and solve for the beta's as I did before? One reason may be that with a large number of features (independent variables) the matrix inversion can be slow, and a computational algorithm like gradient descent may be more efficient.
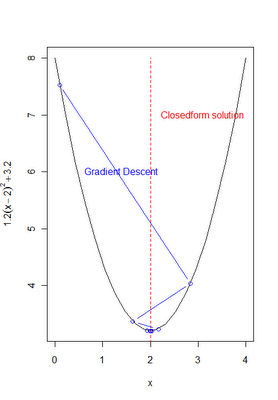
From my earliest gradient descent post, I provided the intuition and notation necessary for performing gradient descent to estimate OLS coefficients. Following the notation from the machine learning course mentioned above, I review the notation again below.
In machine learning, the regression function y = b0 + b1x is referred to as a 'hypothesis' function:
The idea, just like in OLS is to minimize the sum of squared errors, represented as a 'cost function' in the machine learning context:
This is minimized by solving for the values of theta that set the derivative to zero. In some cases this can be done analytically with calculus and a little algebra, but this can also be done (especially when complex functions are involved) via gradient descent. Recall from before, the basic gradient descent algorithm involves a learning rate 'alpha' and an update function that utilizes the 1st derivitive or gradient f'(.). The basic algorithm is as follows:
repeat until convergence {
x := x - α∇F(x)
}
x := x - α∇F(x)
}
(see here for a basic demo using R code)
For the case of OLS, and the cost function J(theta) depicted above, the gradient descent algorithm can be implemented in pseudo-code as follows:
and the gradient descent algorithm becomes:
The following R -Code implements this algorithm and achieves the same results we got from the matrix programming or the canned R or SAS routines before. The concept of 'feature scaling' is introduced, which is a method of standardizing the independent variables or features. This improves the convergence speed of the algorithm. Additional code intuitively investigates the concept of convergence. Ideally, the algorithm should stop once changes in the cost function become small as we update the values in theta.
The following R -Code implements this algorithm and achieves the same results we got from the matrix programming or the canned R or SAS routines before. The concept of 'feature scaling' is introduced, which is a method of standardizing the independent variables or features. This improves the convergence speed of the algorithm. Additional code intuitively investigates the concept of convergence. Ideally, the algorithm should stop once changes in the cost function become small as we update the values in theta.
R-Code:
# ---------------------------------------------------------------------------------- # |PROGRAM NAME: gradient_descent_OLS_R # |DATE: 11/27/11 # |CREATED BY: MATT BOGARD # |PROJECT FILE: # |---------------------------------------------------------------------------------- # | PURPOSE: illustration of gradient descent algorithm applied to OLS # | REFERENCE: adapted from : http://www.cs.colostate.edu/~anderson/cs545/Lectures/week6day2/week6day2.pdf # | and http://www.statalgo.com/2011/10/17/stanford-ml-1-2-gradient-descent/ # --------------------------------------------------------------------------------- # get data rm(list = ls(all = TRUE)) # make sure previous work is clear ls() x0 <- c(1,1,1,1,1) # column of 1's x1 <- c(1,2,3,4,5) # original x-values # create the x- matrix of explanatory variables x <- as.matrix(cbind(x0,x1)) # create the y-matrix of dependent variables y <- as.matrix(c(3,7,5,11,14)) m <- nrow(y) # implement feature scaling x.scaled <- x x.scaled[,2] <- (x[,2] - mean(x[,2]))/sd(x[,2]) # analytical results with matrix algebra solve(t(x)%*%x)%*%t(x)%*%y # w/o feature scaling solve(t(x.scaled)%*%x.scaled)%*%t(x.scaled)%*%y # w/ feature scaling # results using canned lm function match results above summary(lm(y ~ x[, 2])) # w/o feature scaling summary(lm(y ~ x.scaled[, 2])) # w/feature scaling # define the gradient function dJ/dtheata: 1/m * (h(x)-y))*x where h(x) = x*theta # in matrix form this is as follows: grad <- function(x, y, theta) { gradient <- (1/m)* (t(x) %*% ((x %*% t(theta)) - y)) return(t(gradient)) } # define gradient descent update algorithm grad.descent <- function(x, maxit){ theta <- matrix(c(0, 0), nrow=1) # Initialize the parameters alpha = .05 # set learning rate for (i in 1:maxit) { theta <- theta - alpha * grad(x, y, theta) } return(theta) } # results without feature scaling print(grad.descent(x,1000)) # results with feature scaling print(grad.descent(x.scaled,1000)) # ----------------------------------------------------------------------- # cost and convergence intuition # ----------------------------------------------------------------------- # typically we would iterate the algorithm above until the # change in the cost function (as a result of the updated b0 and b1 values) # was extremely small value 'c'. C would be referred to as the set 'convergence' # criteria. If C is not met after a given # of iterations, you can increase the # iterations or change the learning rate 'alpha' to speed up convergence # get results from gradient descent beta <- grad.descent(x,1000) # define the 'hypothesis function' h <- function(xi,b0,b1) { b0 + b1 * xi } # define the cost function cost <- t(mat.or.vec(1,m)) for(i in 1:m) { cost[i,1] <- (1 /(2*m)) * (h(x[i,2],beta[1,1],beta[1,2])- y[i,])^2 } totalCost <- colSums(cost) print(totalCost) # save this as Cost1000 cost1000 <- totalCost # change iterations to 1001 and compute cost1001 beta <- (grad.descent(x,1001)) cost <- t(mat.or.vec(1,m)) for(i in 1:m) { cost[i,1] <- (1 /(2*m)) * (h(x[i,2],beta[1,1],beta[1,2])- y[i,])^2 } cost1001 <- colSums(cost) # does this difference meet your convergence criteria? print(cost1000 - cost1001)